May 4, 2022
Share
Despite its role in disease severity and outcome, the mechanisms behind bone metastases in prostate cancer are not well understood. Rebeca San Martin and her colleagues at the University of Tennessee in the lab of Rachel Patton McCord have connected distinct changes in the 3D genome with prostate cancer progression to metastasis.
The researchers performed chromosome conformation capture (Hi-C) on a cohort of cell lines modeling prostate cancer progression from normal epithelium to bone and distant metastases. Their work conclusively identified several important and distinct changes in the 3D genome, including chromatin compartment identity switches, associated with prostate cancer progression and severity.
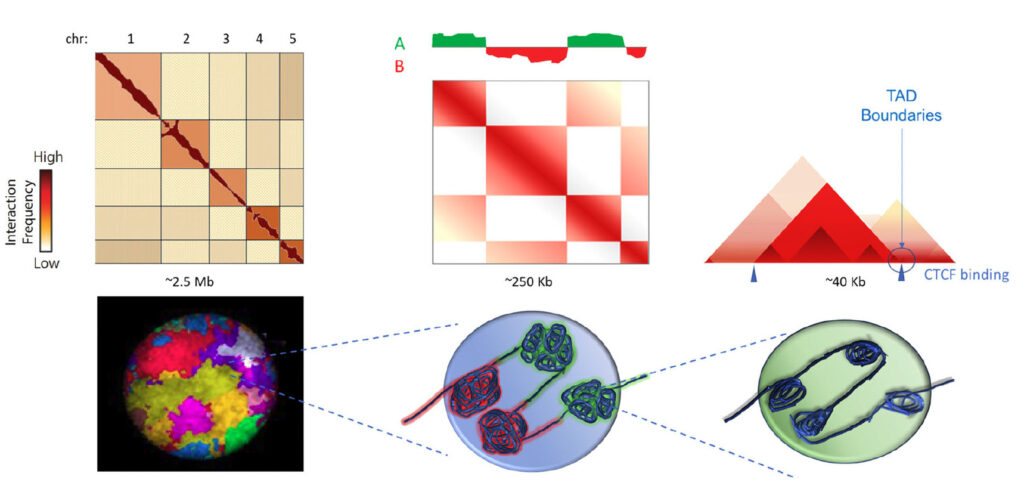
Using 3D genomics with Arima-HiC scientists characterized the hierarchical genome of each of nine cell lines to identify chromosome territories (left), A/B chromosome compartmentalization (center) and topologically associating domains (TADs) within compartments (right).
Prostate cancer is a significant public health concern, being the second most common cause of cancer-related deaths in men. Survival rates for patients with localized disease are nearly 100%, but when detected in the later stages, when metastasis to trabecular bone is common, prostate cancer outcome is poor. Disseminated tumor cells can be detected in the blood of about 25% of patients with localized disease, representing a critical opportunity to identify metastasis biomarkers early in disease.
Several studies have identified mutations, structural alterations, and gene regulatory changes at numerous genomic loci that are associated with a higher risk for prostate cancer. Differences in nuclear lamina content in normal and cancerous prostate cells suggests that changes to the 3D genome might influence cancer-promoting gene expression profiles and the nuclear malleability necessary for cancer cells to metastasize.
Using the Arima Genome-Wide HiC kit, San Martin, Das, Xu, and colleagues conducted a systematic genome architecture comparison on a cohort of nine cell lines modeling prostate cancer progression. Their work, published in the Journal of Cell Biology, suggests that discrete 3D genome structural changes play a deleterious role in prostate cancer progression.
Changes in the 3D Genome Affect Prostate Cancer Progression – Highlights
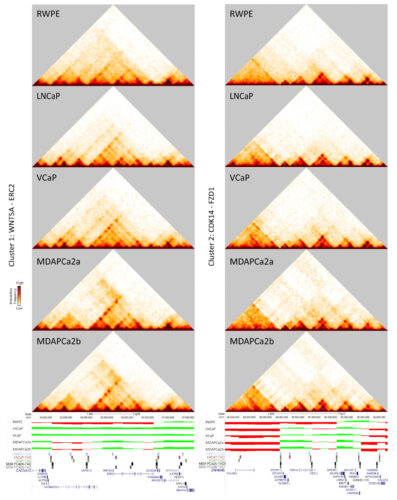
Gene clusters that switch from the B to the A compartment include genes critical for prostate cancer progression. Heat maps, compartment tracks, and TAD boundaries are shown for WNT5A (left) and CDK14 (right), which both change compartments as disease progresses.
- Hi-C identified 48 gene clusters that switch from the B to the A compartment as disease progresses, including androgen receptor, WNT5A, and CDK14. Switches were accompanied by changes in the structure, size, and boundaries of topologically associating domains (TADs).
- NCaP was identified as a central node connecting non transformed epithelium (RWPE) to bone metastatic cell lines (MDAPCa2a/b and VCaP).
- Compartment changes in chromosome 21 were exacerbated with disease progression and may explain, at least in part, the genesis of the TMPRSS2-ERG translocation.
- Gene clusters identified as switching compartments have altered expression in prostate cancer patients, confirming their clinical importance.
The study authors note that results consistent with their own were presented in a 2021 preprint, and therefore propose that “cancer progression is associated with specific changes in the 3D genome structure that arise early in the disease and facilitate an oncogenic expression phenotype.” Furthermore, they hypothesize that “analyzing the 3D genome structure of patient-derived samples could be a prognostic marker for progression and bone metastasis.”
These and other novel insights into the role of 3D genomics in cancer have been identified using Arima Hi-C technology. Discover how in our webinar entitled: Closing in on Undiagnosable Tumors – The Case for 3D Genome Profiling of All Cancers
Learn more about the role of 3D genomics in human health or request a project consultation.
Resources
San Martin et al. (2022) Chromosome compartmentalization alterations in prostate cancer cell lines model disease progression. J. Cell Biology. 2022. 221(2):e202104108; doi: 10.1083/jcb.202104108



