Uncover the Functional and Clinical Significance of Structural Variants
Genomic studies have shown that structural variants – including translocations, inversions, insertions, deletions, and duplications – account for most of the genetic variation between human haplotypes and contribute significantly to disease and disease susceptibility. One of the mechanisms by which structural variants can impact human health is by modifying 3D genome organization and gene regulation; however, this remains largely uncharacterized.
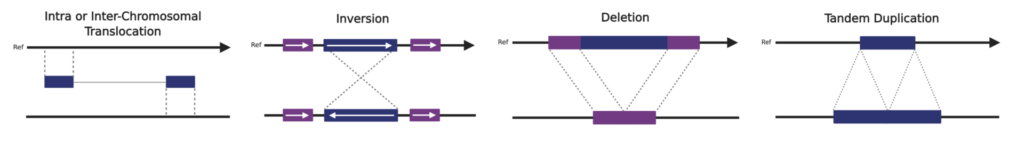
Comprehensive detection of structural variants in both coding and non-coding regions can provide meaningful insights into the impact of those variants on gene function, improve our understanding of disease mechanisms, and identify novel therapeutic targets.
Why Choose Arima Hi-C for Your Research?
Use the proven Arima-HiC+ technology to generate high-quality data from clinically relevant samples
Fast and user-friendly workflows provide efficient sample-to-discovery
Quality you can trust, with built-in QC steps to ensure you get reliable sequencing results every time
Capture more data from hard-to-use samples, including FFPE tissues – regardless of their age
Explore Disease Mechanisms
With Arima Hi-C technology and the Arima-SV bioinformatics pipeline, scientists can now visually and algorithmically identify structural variants in genes known to have clinical significance with high concordance to orthogonal datasets.

A gene fusion was identified from a confirmed fibrosarcoma FFPE sample using the Arima-HiC+ FFPE kit and Arima-SV bioinformatics pipeline. Hi-C heat maps showing relative spatial proximity and breakpoints between chr12 and chr15 resulting in a reciprocal gene fusion between Neurotrophic Receptor Tyrosine Kinase 3 (NTRK3) and ETS Variant Transcription Factor 6 (ETV6) (a). Pictorial representation of the reciprocal NTRK3-ETV6 gene fusion (b).
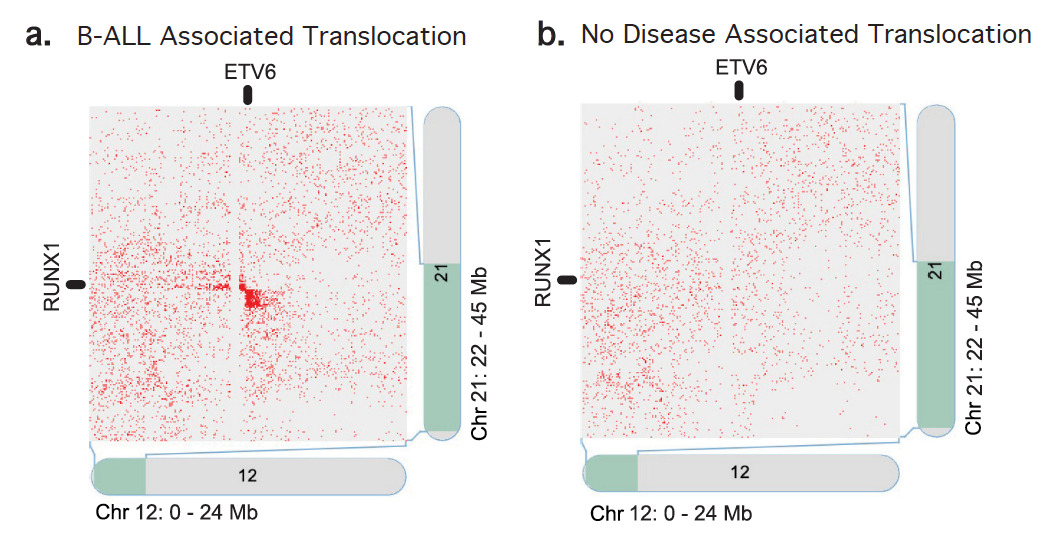
Hi-C contact maps from a peripheral blood sample with a known structural variant for B-ALL, specifically a balanced translocation at the ETV6 locus on chr12 and the RUNX1 locus on chr21 (a). A blood sample with no known variant at that locus demonstrates the absence of abnormal signal in the Hi-C data (b).
Detect Structural Variants to Better Identify Diseases
Identification of the chromosome translocation that drives B-cell acute lymphoblastic leukemia (B-ALL) is typically carried out via invasive bone marrow biopsy. However, with Arima Hi-C technology, driver structural variants were detected in peripheral blood samples. Learn more
Establish New Potential Drug Targets
With Arima-HiC+ for genome-wide HiC scientists were able to identify chromosomal rearrangements associated with ependymoma tumorigenesis – including the formation of new topologically associating domains, known as neo-TADs – in LAMC1 and other genes. By elucidating the regulatory mechanisms underlying the aberrant expression of these genes, they were able to conclude these genes may provide novel therapeutic targets. Learn more
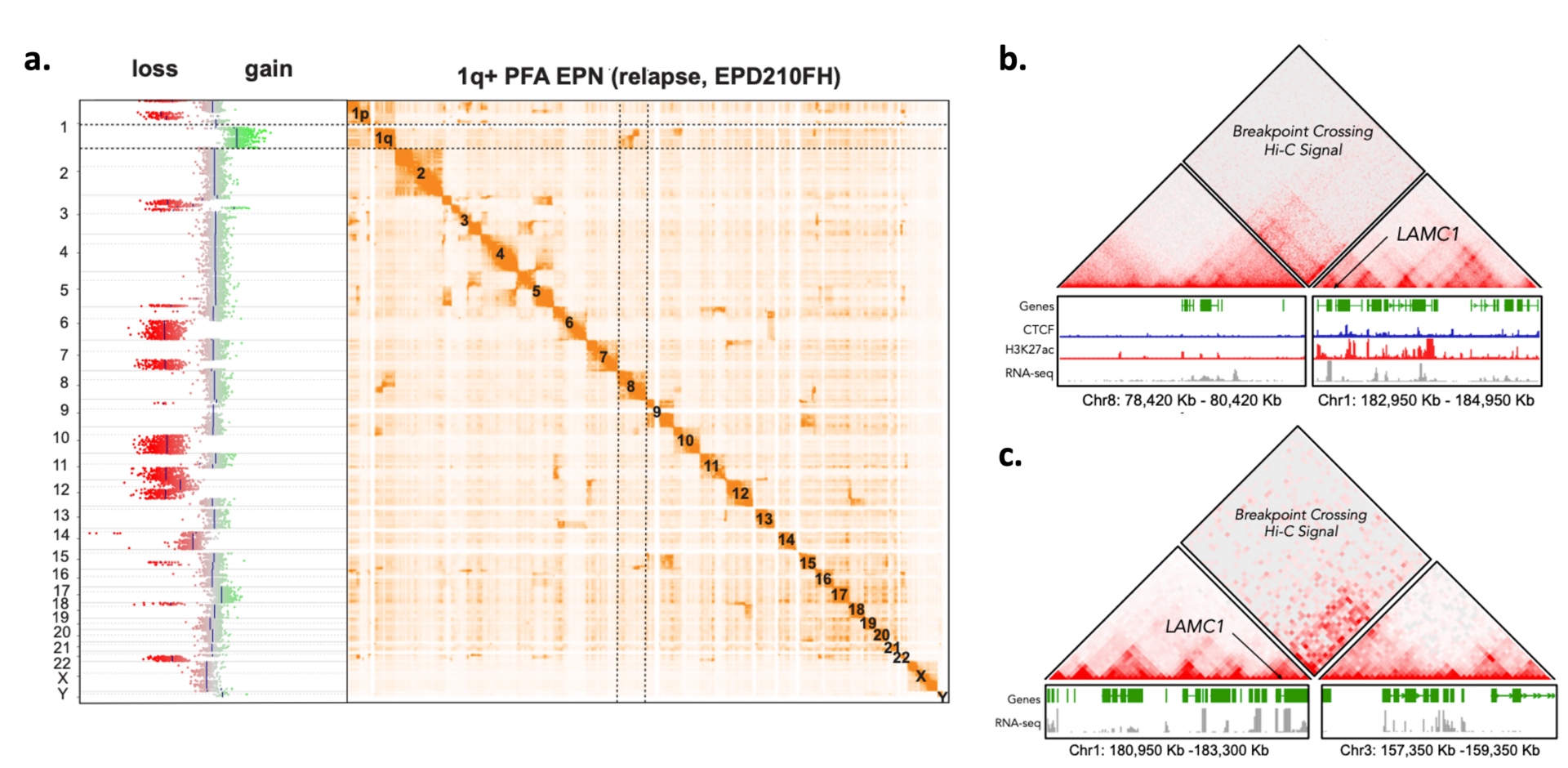
Genome-wide Hi-C heat maps were used to identify complex inter-chromosomal structural variants, including an inversion that involves chr1q and chr8 (a), resulting in the formation of a neo-TAD in cultured cells that alters the regulatory environment of the LAMC1 gene which activates gene transcription (b,c).
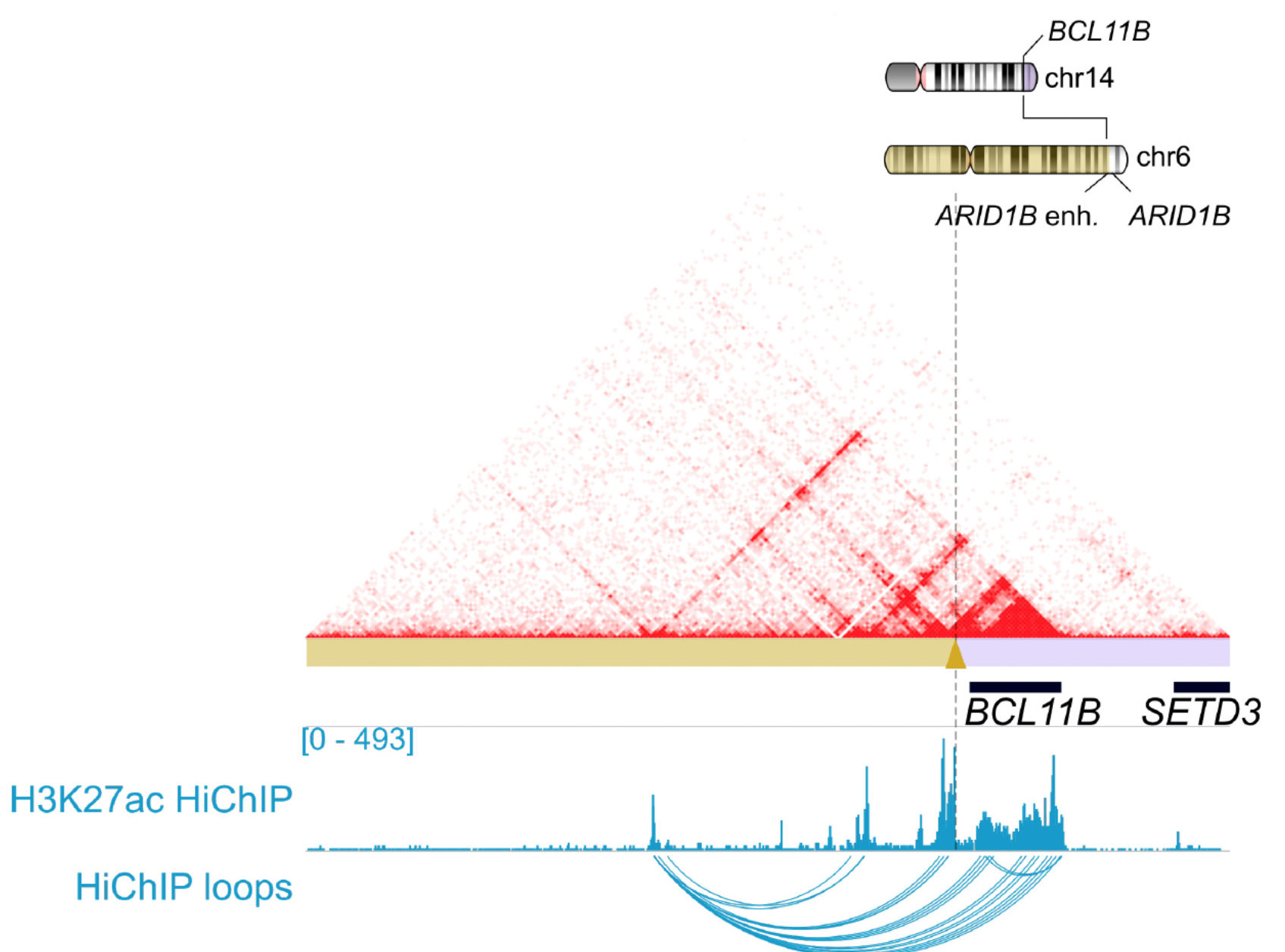
HiChIP was used to identify that BCL11B rearrangements rewire CD34+ hematopoietic stem and progenitor cell super-enhancers. This demonstrated that a novel enhancer hijacking mechanism drives oncogenic BCL11B expression in lineage ambiguous leukemia.
Link Structural Variants to Impacts on Gene Function
To understand the role of structural variants in tumorigenesis it is critical to know how they impact gene regulation. With Arima Hi-C technologies, you can capture the location of structural variants, gene activity with H32K27ac, and the resulting 3D interactions caused by the structural variants to determine the impacts on gene regulation. Learn more
Matija Snuderl, MD
Director of Molecular Pathology and Diagnostics, NYU Langone Medical CenterHiC+ FPPE
The Arima-HiC+ FFPE Kit and workflow are ideal for the detection and discovery of structural variants from archival or clinical samples.
Learn More