Reveal Novel Cancer Biomarkers and Therapeutic Targets
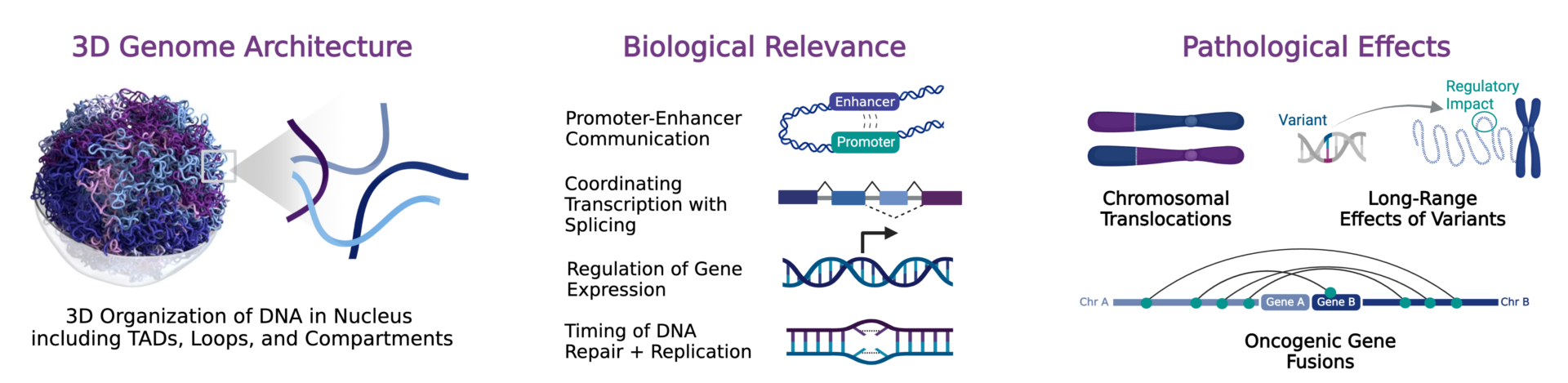
Gain a complete understanding of the regulatory landscape of cancer genomes with 3D genomic information to reveal the pathological implications of these alterations, including chromosome translocations, the creation of oncogenic fusion transcripts, and the long-range effects of variants on gene regulation. Use 3D genomic signatures to discover and validate novel diagnostic or prognostic cancer biomarkers, identify therapeutic targets, or assess therapeutic response to treatment.
Why Choose Arima Technology for Your Cancer Sequencing?
Discover and detect novel gene fusions, including those that are missed by other technologies
See how 3D genome architecture can elucidate disease mechanisms revealing new cancer biomarkers and targets
Discover What’s Possible with 3D Cancer Genomics
Detect Clinically Relevant Gene Fusions
Learn how 3D genomics can add diagnostic value by identifying novel molecular drivers and potential therapeutic targets in tumors with no previously detectable drivers.
Watch to see how after comprehensive DNA and RNA sequencing failed to produce a causative driver in a case of pediatric glioblastoma, 3D genomics using Arima technology enabled the identification of the causative PD-L1 rearrangement.
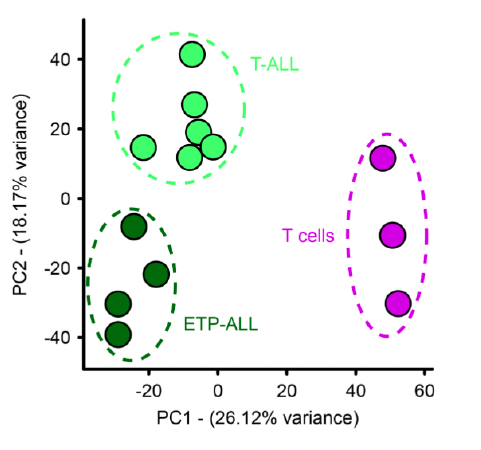
Arima Hi-C analysis identified genome-wide 3D chromatin differences between normal T cells and T-ALL subtypes.
Explore Disease Mechanisms
3D genomics yielded novel insights into the disease mechanisms of T-cell acute lymphoblastic leukemia (T-ALL). Scientists discovered that the 3D organization of an oncogene relative to its enhancer could affect the progression and treatment of T-ALL.
Kloetgen, A., et al. (2020). Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nature Genetics.
Identify Enhancer Hijacking
3D genomics demonstrated that an enhancer hijacking mechanism drives oncogenic BCL11B expression in lineage ambiguous leukemia.
Montefiori, L. et al. (2021) Enhancer hijacking drives oncogenic BCL11B expression in lineage ambiguous stem cell leukemia. Cancer Discovery.
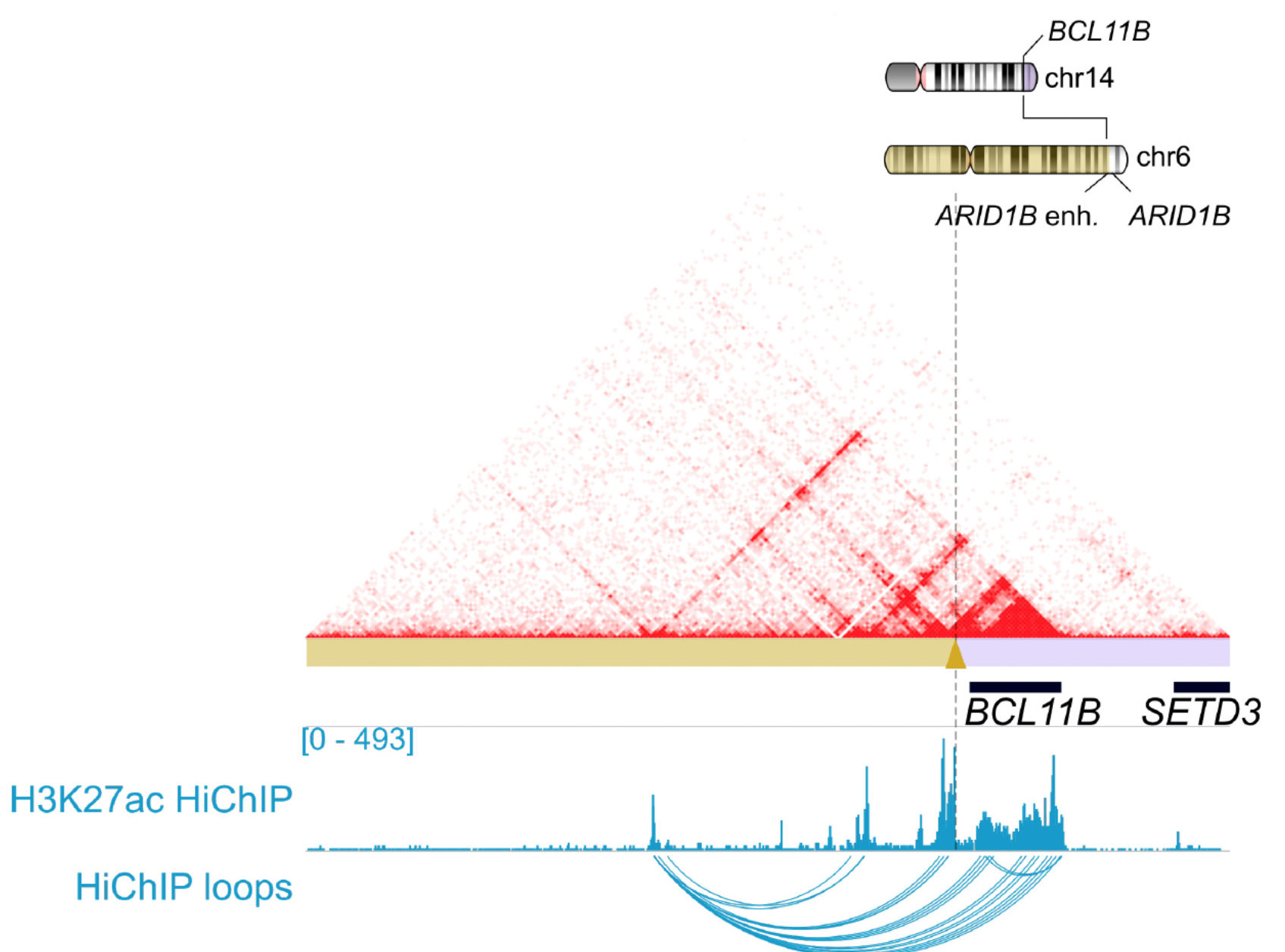
HiChIP analysis showed that structural rearrangements result in aberrant gene-promoter interactions that generate de novo super-enhancers through a high copy tandem amplification event downstream of BCL11B.
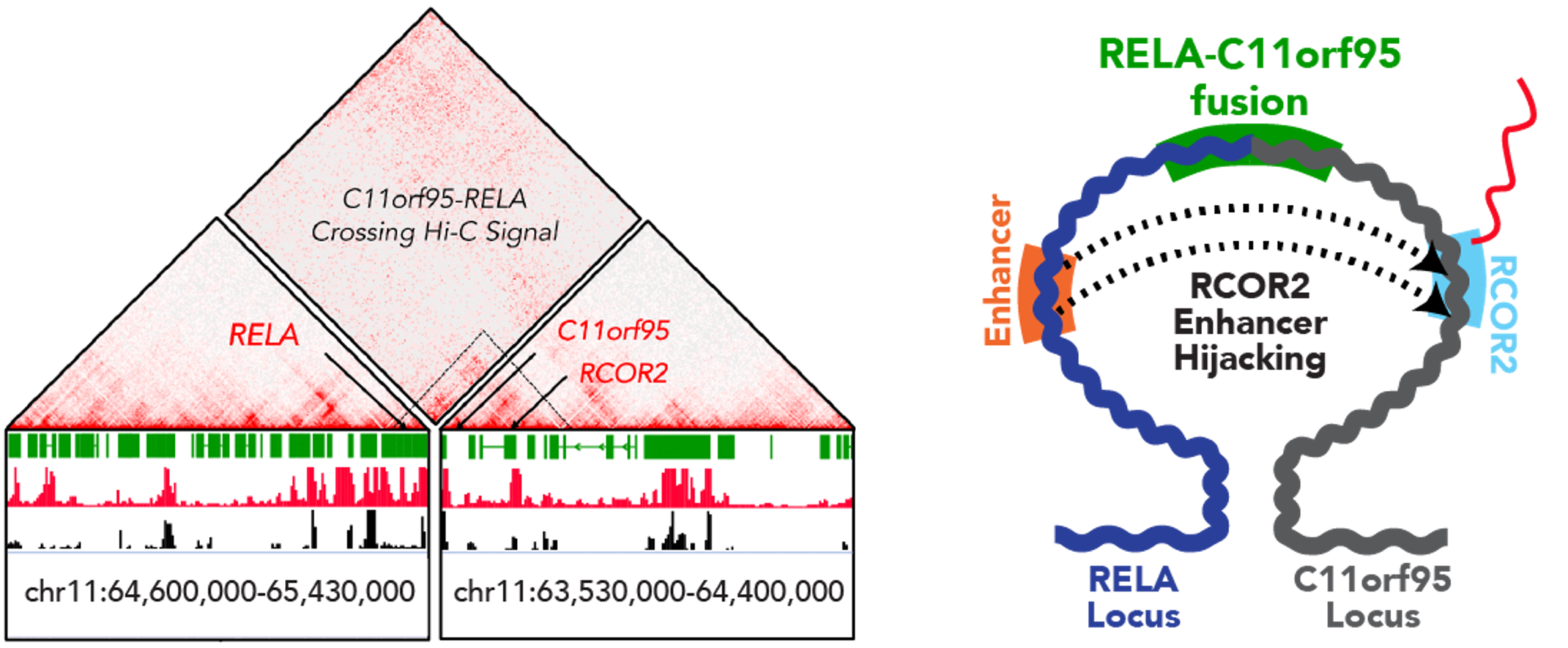
Develop Novel Therapeutic Approaches
3D genomic insights were gained from FFPE samples, revealing recurrent chromosomal rearrangements around LAMC1 and other genes. This enabled the identification of the regulatory mechanisms underlying the aberrant expression of these genes and putative therapeutic targets for ependymoma tumorigenesis.
Okonechnikov, K. et al. (2020). Oncogenic 3D genome conformations identify novel therapeutic targets in ependymoma. Preprint.
Two Ways To Get Started with Cancer Genome Sequencing
Arima Kits
Use our sample and library preparation kits to advance your understanding of cancer genomics
Arima Services
Let our scientists share their expertise in sample prep, library construction, and bioinformatics