March 28, 2022
Share
Understanding non-coding genetic variants and their functional role in human disease remain a significant challenge. By combining their decades of experience in pharmacogenetics and epigenetics, scientists from the Mayo Clinic, Richard Weinshilboum and Tamas Ordog, have made significant gains toward elucidating the functional roles of several non-coding variants.
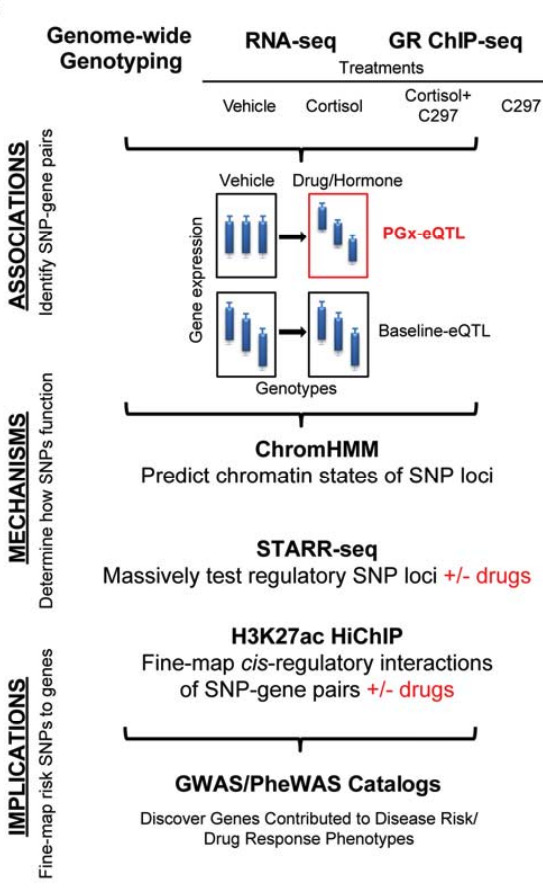
Overview of study design in which H3K27ac HiChIP was used to capture chromatin conformation of enhancer-promoter and enhancer-enhancer interactions in a high-resolution manner before and after drug treatments.
Building upon previous work where they identified genetic variants that influence gene expression only after exposure to a hormone or drug, the doctors and their research colleagues used glucocorticoid signaling as a model system to demonstrate in a genome-wide manner that glucocorticoid exposure can trigger disease risk variants to influence the expression of genes involved in autoimmunity, metabolic and mood disorders, osteoporosis, and cancer.
Glucocorticoids are potent and indispensable anti-inflammatory and immunosuppressive medications that maintain various metabolic and homeostatic functions necessary for life. They signal through the glucocorticoid receptor (GR), which is present in almost every tissue of the human body and has been under intense study for decades. GR signaling plays a role in several major human diseases and is, therefore, an important focus of research connecting novel gene-environment associations and disease.
To better understand non-coding genetic variants and their role in disease, many are turning to expression quantitative trait loci (eQTLs). eQTLs are identified by associating non-coding variants with gene transcription. They are often dynamic in nature, impacted by pathogens, cellular differentiation, or drugs/hormones, and have proven a powerful tool for explaining novel functions of non-coding variants and for providing insight into the molecular mechanisms underlying gene-environment interactions, including gene-drug interactions.
Using Arima HiChIP library preparation and sequencing, Weinshilboum, Ordog, and their research team systematically fine-mapped genotype-phenotype interactions during glucocorticoid exposure, advancing the fine-mapping of disease risk and pharmacogenomic loci. They detail their work in this bioRxiv preprint.
Using 3D Genomic to Understand Non-Coding Variants – Highlights
- Glucocorticoids “unmasked” loop-induced functional transcriptional regulation activity of several PGx-eQTLs.
- The current study helped add clarity to the functional mechanisms of several previously recognized eQTLs. For example, a mechanistically unexplained variant that had been associated with breast cancer risk was a cortisol-dependent PGx-eQTL for the Microtubule Associated Serine/Threonine Kinase Family Member 4 (MAST4) gene. Repression of MAST4 expression in the current study was due to a GR-responsive intergenic enhancer looping to MAST4 and transcriptionally regulating it. Repression was reversed after antagonist treatment.
- A total of 30 disease risk loci were found to behave as PGx-eQTLs, spanning a range of conditions including several immune and autoimmune-related diseases, mood disorders, osteoporosis, and cancer.
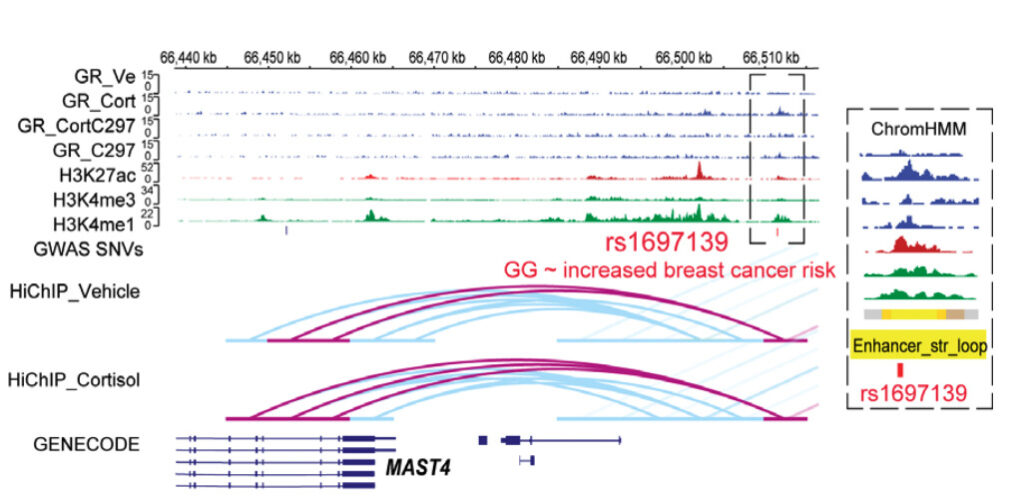
Example of glucocorticoid receptor-dependent pharmacogenomic (PGx)-eQTLs with functional implications for disease risk. Figure shows a SNP, in a genotype-dependent fashion, modulating a GR-responsive intergenic enhancer that interacts across 40,000 base pairs to MAST4, thereby transcriptionally regulating that gene.
The authors note that “by systematically fine-mapping genotype-phenotype interactions in which measurable environmental factors such as drug or hormone exposure were taken into account, we uncovered potential novel risk genes for a range of diseases in which the pharmacological or physiological stimuli played important roles. The function of these SNPs was usually “masked” in the absence of exposure to hormones or related drugs. As a result, this study has added a novel perspective to functional genomics by providing a mechanistic framework for additional studies of ligand-dependent “silent” non-coding genetic variants to advance the fine-mapping of disease risk and pharmacogenomic loci.”
These and other novel insights into transcriptional regulation associated with the 3D structure of the genome as a major factor in disease mechanisms have been identified using Arima Hi-C technology.
Learn more about the role of 3D genomics in human health or request a project consultation.
Resources
Le Nguyen, T. T., Gao, H., Liu, D., Ye, Z., Lee, J.-H., Shi, G., Copenhaver, K., Zhang, L., Wei, L., Yu, J., Zhang, C., Li, H., Wang, L., Ordog, T., & Weinshilboum, R. M. (2021). Glucocorticoids Unmask Silent Non-Coding Genetic Risk Variants for Common Diseases. BioRxiv Preprint. https://doi.org/10.1101/2021.12.01.470787



