January 30, 2023
Share
What is Multi-Omics?
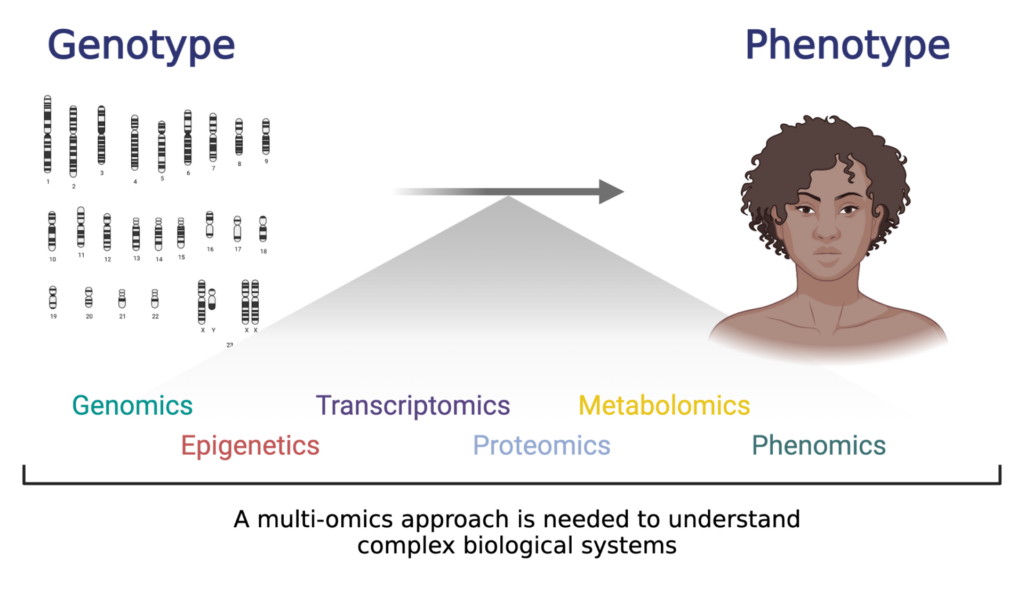
Multi-omics seeks to bridge the gap between genotype and phenotype in complex biological systems.
Multi-omics involves integrating various levels of data, including genomic, transcriptomic, epigenomic, proteomic, and metabolomic, to understand complex biological systems. Ultimately, any multi-omics project aims to bridge the immense biological gap between genotype and phenotype.
Since its advent, the most established omic, genomics, has revolutionized biomedical research and our understanding of the natural world. However, it is clear using a single omic technology is no longer sufficient to understand complex biological systems truly. Multi-omics is already emerging as arguably the most important modern technique in scientific research by offering the opportunity to expand our understanding of biology using integrated system-level approaches exponentially.
What is 3D Genomics?
3D genomics involves exploring the 3-dimensional organization of DNA in the nucleus to reveal insights into the genome’s sequence, structure, and regulatory landscape. Although techniques to understand the 3D organization of chromosomes were first described in the rat prolactin gene by Cullen et al. (1993) and then in yeast by Dekker et al. (2002), the field of 3D genomics is relatively nascent. It is now known that there are multiple levels of chromatin organization in the nucleus of cells, including chromosomal territories, compartments, domains, and looping structures that coordinate regulatory interactions.
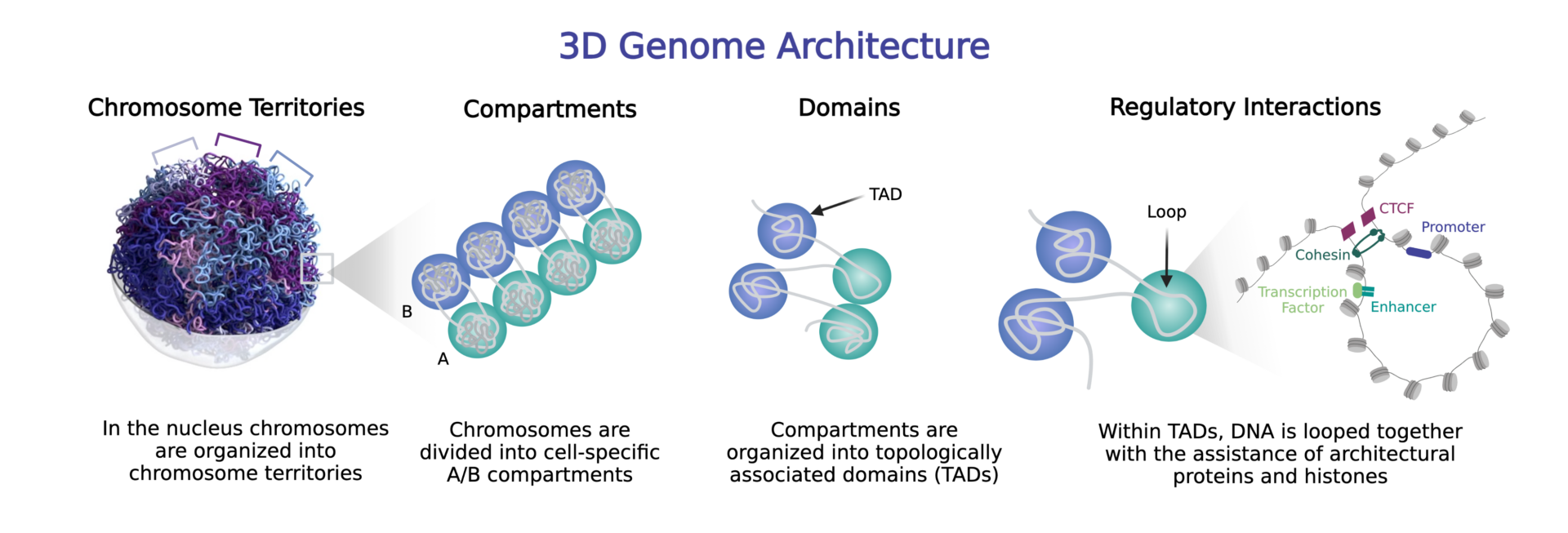
The 3D genome is organized at multiple levels into chromosome territories, compartments, domains including topologically associated domains (TADs), and looping structures.
Many papers have described 3D genome organization and its impacts on cellular function, development, disease, and immune function; however, we are just beginning to elucidate the breadth of the 3D genome’s impacts on biological function.
Where Does 3D Genomics Fit in the Multi-Omics Landscape?
3D genomics, as the name suggests, sits next to genomics in the multi-omics landscape. Unlike genomics, the breadth of information that 3D genomics provides extends far beyond the linear sequence of DNA. Also adjacent to 3D genomics is epigenomics, the study of non-sequence modifications that affect chromatin state. Therefore, as a field of science, 3D genomics sits between genomics and epigenomics, serving as a valuable connector within the multi-omics landscape.
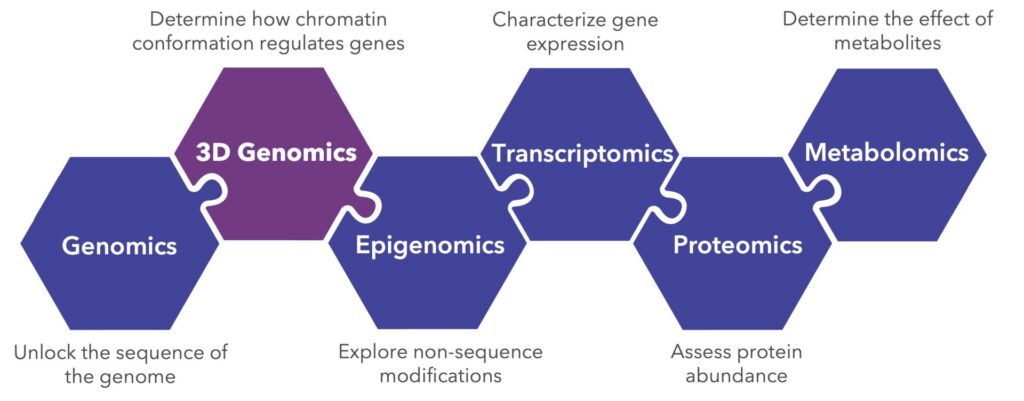
3D genomics, part of the multi-omics landscape, occupies the space between genomics, which looks at the linear sequence of DNA, and epigenomics, which explores non-sequence-based modifications to DNA.
In 2016, Tordini et al. highlighted 3D genome conformation as a critical integrator of multi-omics data in the realm of cancer research. The authors of this paper keenly projected that multi-omics analysis with the inclusion of chromatin conformation data, will broadly enhance our understanding of the biology of cancer, leading to the discovery of novel diagnostic, prognostic, and therapeutic approaches that will ultimately improve the disease outcomes.
To learn more about the role of 3D genomics in diverse areas of biology, explore these review articles:
- Dubois, F., et al. (2022). Structural variations in cancer and the 3D genome. Nature Reviews Cancer.
- Cuartero, S., et al. (2022). Three-dimensional genome organization in immune cell fate and function. Nature Reviews Immunology.
- Boltsis, I., et al. (2021). Chromatin Conformation in Development and Disease. Frontiers in Cell and Developmental Biology.
- Dong, X., et al. (2021). Review of multi-omics data resources and integrative analysis for human brain disorders. Briefings in Functional Genomics.
Incorporating 3D Genomics into a Multi-Omics Approach
What information does 3D genomics provide that makes it such a valuable tool in the multi-omics toolkit? The utility of chromatin conformation data is quite varied. It can be used to assemble chromosome-scale genomes, link non-coding disease-associated risk variants to target genes, understand the mechanisms of gene regulation, and detect gene fusions missed by other technologies, to name a few. 3D genomics data is incredibly rich and provides basic, translational, and clinical researchers with novel biological insights that no other omic offers.

3D genomic data offers a wealth of information and provides basic, translational, and clinical researchers with novel biological insights that no other omic offers.
Exploring Gene Regulation with Multi-Omics
One of the most well-established uses of 3D genomic data is understanding gene regulation in order to explore mechanisms of cellular or organismal development, understand disease processes, or map cell-type specific functions.
Because every cell in an organism contains a copy of the same genome, transcriptional regulation is essential to the diverse function observed across cell type, time, disease states, the environment, genetic mutations, and many more factors. To explore how any one of these variables impacts gene expression, scientists frequently use transcriptomics to detect differential expression of genes. This compelling technique has significantly added to our understanding of transcriptional regulation. Still, RNA sequencing does not provide any information on what is causing that altered gene expression. And that is where 3D genomics can truly add value as part of a multi-omics approach.
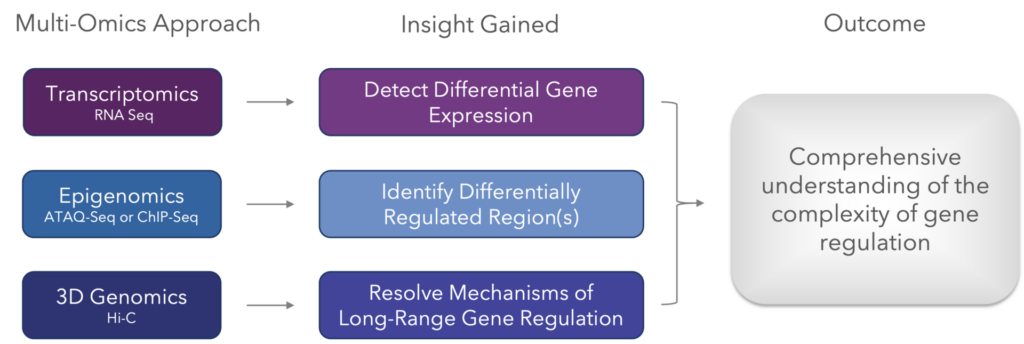
The common combination of multi-omic techniques used generate a comprehensive understanding of gene regulation.
By incorporating 3D genomic information, it is possible to identify what is causing the differential expression observed by transcriptomic data. Below is an example multi-omics study that can provide substantial information into the expression of oncogenes between cancer and normal cells. Briefly, when looking to compare two populations of cells (eg: cancer vs normal cells), using a combination of three approaches, namely transcriptomics (RNA-Seq data), 3D genomics (chromatin conformation Hi-C data) and epigenomics (ChIP-Seq and/or ATAC-Seq data) is very powerful.
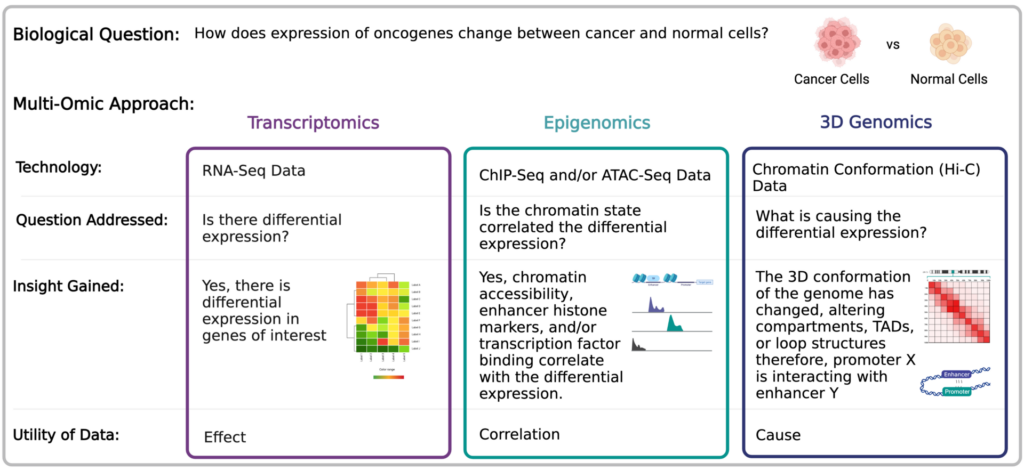
The combination of transcriptomic, epigenomic, and 3D genomic data can provide a comprehensive understanding of gene regulation.
The RNA-seq data addresses the question is there differential expression between these two samples? This yields baseline data that serves as the effect but does not provide information on the cause of the altered expression. Epigenomics data then provides an additional layer of correlative evidence that indicates how the chromatin state is contributing to altered expression levels of genes of interest. Finally, chromatin conformation data is what provides information on what is causing that altered expression, for instance, that promoter of oncogene X being brought into proximity of enhancer Y.
Research Demonstrating 3D Genomics Adds Significant Value to Multi-Omics
There are many published examples in the Arima Publication Library, but below are some key publications that highlight the value of 3D genomics as part of a multi-omics approach:
- Kloetgen, et al. (2020) demonstrated in a Nature Genetics paper that the 3D organization of an oncogene relative to its enhancer could affect the progression of T-cell acute lymphoblastic leukemia.
- Orozco, (2022) combined GWAS data and 3D genomics to understand the role of disease-associated variants in non-coding regions in autoimmune disorders. Check out the webinar describing this work.
- Marchal, et al. (2022) used 3D genomics, GWAS data, and epigenetic information to explore high-resolution genome topology of the human retina, which uncovered super enhancer-promoter interactions at tissue-specific and multifactorial disease loci.
- Lopes, et al. (2021) coupled CRISPR, single-cell RNA sequencing, and 3D genomics to reveal novel non-coding regulatory elements that contribute to oncogenic transcriptional programming and cell proliferation of ER+ breast cancer cells. Read the case study about this research.
- Luo, et al. (2022) used joint single-nucleus multi-omic profiling, including sn-m3C-Seq, to reconstruct the regulatory hierarchy for 63 human cortical cell populations.
Conclusions
3D genomics is an emerging but essential part of any research into the mechanisms of gene regulation and beyond. It is most informative when incorporated as part of a multi-omics approach and has been demonstrated to yield significant biological insights across many areas of research.



