June 21, 2022
Share
Gene transcription occurs within the context of a 3-dimensional chromatin architecture, yet little is known about how this architecture is reorganized in various cancers. In the laboratory of Daeyoup Lee at Korea Advanced Institute of Science and Technology, Taemook Kim, Sungwook Han, Yujin Chun, and colleagues recently charted the 3D genomic architecture of triple-negative breast cancer (TNBC).
The researchers used in situ Hi-C to compare 3D genomic features in TNBC cell lines and tissues versus normal cell lines and tissues. They found dramatic changes in 3D chromatin structures — including compartment domains, TADs, and chromatin loops — that underlied the altered epigenetic and transcriptomic features of TNBC cells.
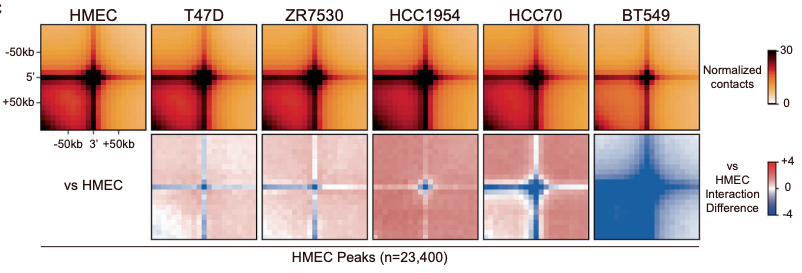
Aggregate peak analysis showing more dramatic loss of Hi-C interactions (chromatin contacts) in the TNBC cell line BT549 than other breast cancer cell lines (T47D, ZR7530, HCC1954, and HCC70) when compared to the normal HMEC cell line.
About 1 in 5 breast cancers are triple-negative (lacking estrogen receptor [ER], progesterone receptor [PR], and human epidermal growth factor receptor 2 [HER2] expression). In addition to being the most aggressive form of breast cancer, TNBC exhibits high heterogeneity across invasive tumors and is not well addressed by conventional treatments such as endocrine therapy and chemotherapy. Illuminating the 3D architecture of these cells could supply important insights to improve the diagnosis and treatment of TNBC.
Daeyoup Lee’s team leveraged the Arima Genome-Wide HiC Kit to analyze the 3D genomes of normal and TNBC cell lines and tissues using Hi-C technology. Their findings, which they published in Experimental & Molecular Medicine, provide the first surveys of the 3D genomes of breast cancer.
Using 3D Genomics to Understand Triple-Negative Breast Cancer — Highlights
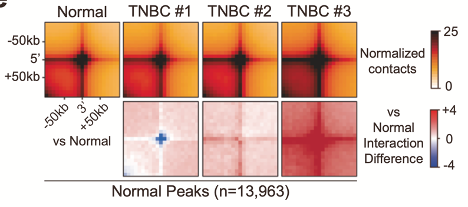
Aggregate peak analysis showing heterogeneous changes in Hi-C interactions (chromatin contacts) across 3 TNBC tissues compared to normal tissue.
- The TNBC cell lines BT549 and HCC70 exhibited more dramatic deviations from normal (HMEC) 3D chromatin structures — including compartment domains, topologically associating domains (TADs), and chromatin loops — than luminal A, luminal B, and HER2+ breast cancer cell lines
- Compartment domain switches between BT549 and HMEC cells were associated with changes in H3K27ac enrichment, chromatin accessibility (ATAC-seq), and the expression of genes related to basal carcinoma, cell cycle, and p53 signaling pathways
- Local 3D chromatin structures identified in BT549 cells are only partially conserved (31% of weakened/strengthened TADs) in three TNBC tissues, presumably due to heterogeneity across TNBC tissues
- Due to TNBC heterogeneity, loss of normal 3D chromatin structures may be better targets for future diagnostic and therapeutic efforts
Although the researchers identified TNBC-specific chromatin loops, they cautioned that, because of the heterogeneity across TNBC tumors, “one can speculate that as we increase the number of examined TNBC patients (or TNBC tissues), certain TNBC-specific 3D chromatin structures may be stochastically detected.” Because of this, they propose that “normal-specific chromatin loops, which are weakened or barely detectable in TNBC tissues, may be a more efficient target feature for diagnostic and therapeutic development in TNBC.”
Learn more about 3D genomics for cancer research or request a project consultation.
Resources
Kim, T., Han, S., Chun, Y., Yang, H., Min, H., Jeon, S. Y., Kim, J. I., Moon, H. G., & Lee, D. (2022). Comparative characterization of 3D chromatin organization in triple-negative breast cancers. Experimental & Molecular Medicine, 10.1038/s12276-022-00768-2.



