December 14, 2022
Share
Single-cell 3D genomics is an emerging approach that combines the resolution of single-cell technology with the power of 3D genomics to reveal the genome’s sequence, structure, and regulatory landscape. In this getting started guide, you will find answers to common questions and resources to get you started with single-cell 3D genomics.
What is Single-Cell 3D Genomics?
All cells in an individual carry the same DNA sequence, yet their morphology and function in the context of cell types, tissues, and organs are vastly different. This diversity is achieved by cells expressing different sets of genes. 3D genomics uses chromatin conformation capture to characterize how genome organization in the nucleus impacts gene regulation, including through gene-enhancer interactions. By coupling 3D genomics with single-cell sequencing, an extremely high-resolution view of how genome organization affects gene expression across cell types can be uncovered.
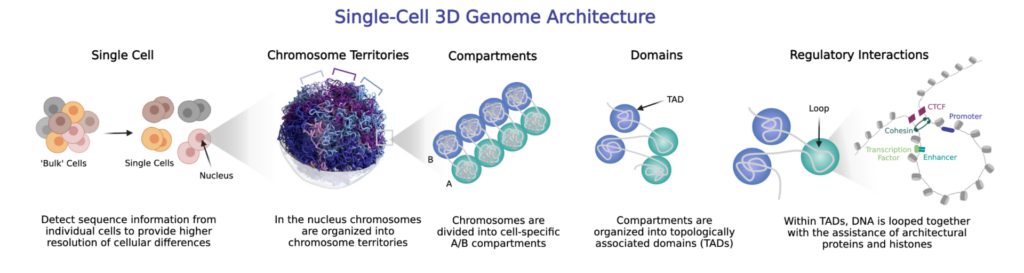
Why use a Single-Cell 3D Genomics Approach?
3D genomics can be incredibly powerful in “bulk” assays; however, many diseases manifest in distinct cell subtypes or disease microenvironments. To accurately develop mechanistic models of cellular function, differentiation, and diversity to elucidate the basis of their genotype–phenotype connections, it is critical to use a single-cell approach. Single-Cell 3D Genomics Approaches are particularly useful for understanding the diverse cell types in the brain. Learn more: The Brain: A Stimulating Frontier for 3D Genomic Insight
What is the Ideal Experimental Design for Single-Cell 3D Genomics?
Like with so many approaches, 3D genomic data is most useful when incorporated as part of a multi-omic approach. Here are the techniques that, when combined, offer the most valuable biological insights for understanding the mechanisms of gene regulation.
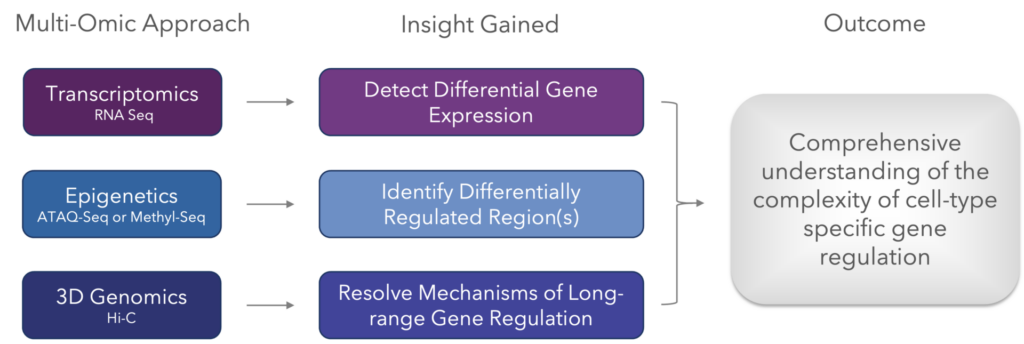
What Methods Exist for Single-Cell 3D Genomics?
Two primary methods have been developed for coupling 3D genomics and single-cell data:
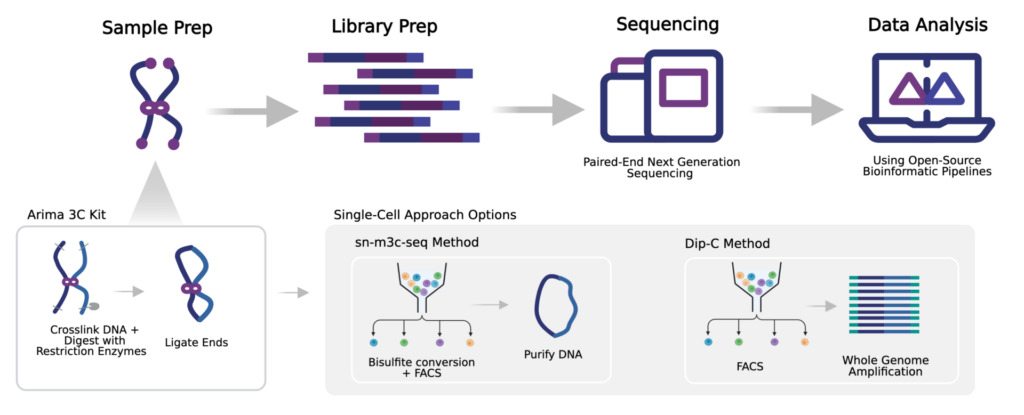
Single-nucleus methyl-3C sequencing (sn-m3C-Seq)
This approach, pioneered by scientists at UCLA and the Salk Institute, can be used to separate heterogeneous cell types robustly. Features of this method include:
- Provides multi-modal single-cell data that is automation-friendly and produces data that correlates well to its bulk analog
- Delineates cell types using single-cell methylomes, then analyze 3D chromatin of each cell type
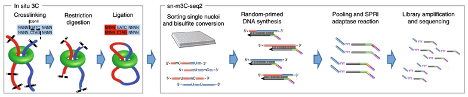
Lee DS, et al (2019). Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat Methods.16(10):999-1006.
Diploid Chromatin Conformation Capture (Dip-C)
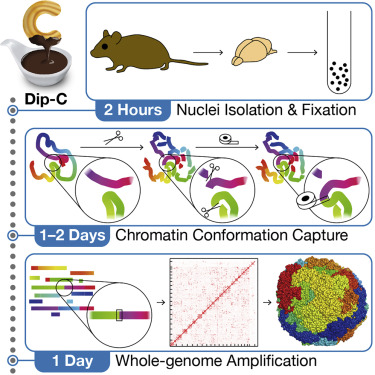
This approach was developed by scientists at Stanford University and is a robust, fast, low-cost method that does not require liquid handlers. Features of this method include:
- Provides the highest number of contacts per cell (~25k) but requires 50-100K contacts per cell to distinguish sub-types
- Facilitates 3D structural modeling and cell-type delineation from 3D “structure-types”
Tan, L. (2021). Determining the 3D genome structure of a single mammalian cell with Dip-C. STAR Protocols, Volume 2, Issue 3, 17.
How Does Arima Technology Interface with Single-Cell Approaches?
Currently, we offer the Arima 3C Kit, which enables DNA crosslinking, enzymatic digestion, and ligation of crosslinked DNA. The product of these reactions then feeds into either the sn-m3C-Seq or Dip-C approaches, to be followed by sequencing and data analysis.
How Can You Learn More about Single-Cell 3D Genomics?
Watch our webinar featuring leading researchers from Stanford, UCLA, and the Salk Institute, who are using single-cell 3D genomic approaches to understand development, disease, and aging in the human brain.
Additionally, several new and exciting research publications feature single-cell 3d genomic approaches to resolve complex biology. Here are some of our favorites:
- Liu, H., et al. (2021). DNA methylation atlas of the mouse brain at single-cell resolution. Nature 598, 120–128
- Tan, et al. (2021). Changes in genome architecture and transcriptional dynamics progress independently of sensory experience during post-natal brain development. Cell, Volume 184, Issue 3, Pages 741-758.e17
- Heffel, M. G., et al., (2022). Epigenomic and chromosomal architectural reconfiguration in developing human frontal cortex and hippocampus. bioRxiv Preprint.
- Tian, W., et al. (2022). Epigenomic complexity of the human brain revealed by single-cell DNA methylomes and 3D genome structures. bioRxiv Preprint.
Ready to Get Started?
Connect with an Arima scientist to get started with single-cell 3D genomics.



