October 20, 2022
Share
To say we at Arima always have ‘3D genomics on the brain’ would be an understatement. But never has it been more true than with the recent announcement of a five-year, $126 million NIH grant to a team led by scientists at the Salk Institute for Biological Sciences, which will enable the creation of a new Center for Multiomic Human Brain Cell Atlas. As part of the NIH Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative, the new Center will endeavor to map the human brain on a cellular level to understand how brains operate and age.

As part of this vast endeavor and approach, Arima technology will be utilized towards exploring the 3D genome of the brain at the single-cell level. This will allow the research team to map chromatin conformation and its effects on gene regulation across the vast diversity of brain cell types.
“We are just starting to understand the breadth of the role 3D genomics plays in human brain function and aging. Through the Center for Multiomic Human Brain Cell Atlas, we aim to elucidate how 3D genomics, epigenomics, and transcriptomics interplay in the brain to ultimately reveal new paradigms for understanding how pathological changes in particular groups of brain cells could cause neurologic and neuropsychiatric disorders.”
– Joseph Ecker, Director of the Genomic Analysis Laboratory, Salk Institute
To celebrate this exciting time in the 3D genomics of the brain, here is a look at some of the fascinating areas of neuroscience that are being explored with Arima technology.
Single-cell 3D Genomics of the Brain
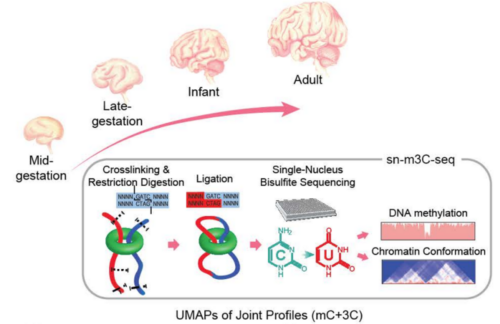
Study design using single-nucleus profiles of chromatin conformation and DNA methylation (sn-m3C-seq).
In a newly published biorxiv preprint from a significant collaborative effort led by Chongyuan Luo of UCLA, in partnership with scientists from the Salk Institute and UCSF, the explored epigenomic and chromosomal architectural reconfiguration in the developing human frontal cortex and hippocampus.
Using more than 53,000 joint single-nucleus profiles of chromatin conformation and DNA methylation (sn-m3C-seq), they showed that the remodeling of DNA methylation predominantly occurs during late-gestational to early infant development and is temporally separated from chromatin conformation dynamics and that neurons, glial cells, and non-brain tissues have varied domain or compartment dominated chromatin conformations. Moreover, this work reconstructed the regulatory programs of cell-type differentiation and found putatively causal common variants for schizophrenia strongly overlap with chromatin loop-connected, cell-type-specific regulatory regions.
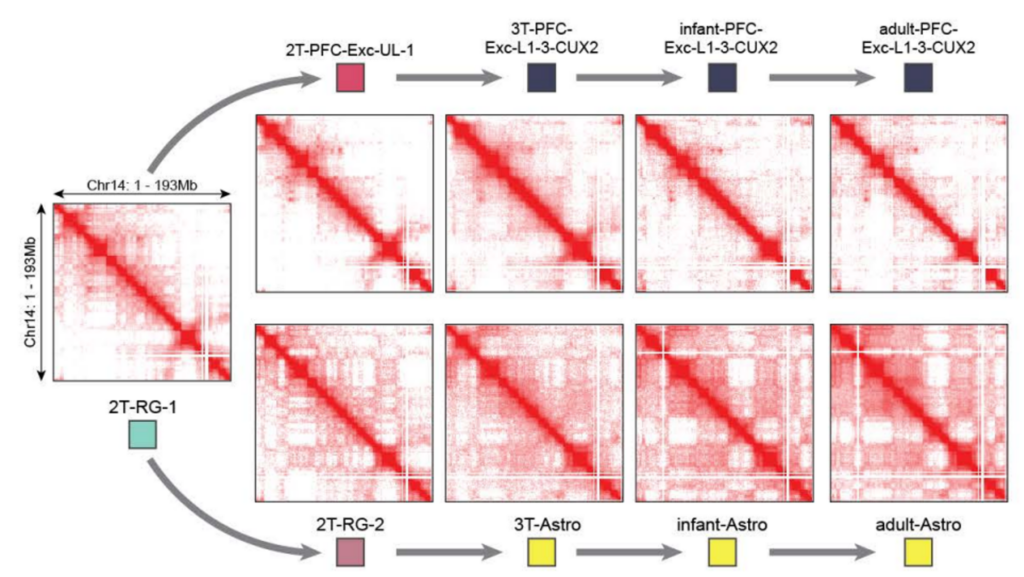
Chromatin interaction profiles of developing neuron and astrocyte cell populations.
“Single-cell 3D-regulome profiling is a powerful, multi-dimensional approach that reveals gene regulatory dynamics and genomic structural rearrangements. We anticipate the approach will enable many discoveries in developmental biology, human genetics, and cancer biology and lead to new biomarkers and improved disease treatment strategies.”
– Chongyuan Luo, Assistant Professor, Human Genetics, UCLA
Heffel, M. G., et al., (2022). Epigenomic and chromosomal architectural reconfiguration in developing human frontal cortex and hippocampus. bioRxiv Preprint.
3D Genomics in Psychiatric Disease Risk
Early seminal work in psychiatric disease risk variants explored cell type-specific chromosomal conformations associated with schizophrenia risk loci. This work, published in Science, showed that neural differentiation is associated with higher cell-type-specific 3D genome remodeling and spatial expansion. Specifically, developmentally regulated chromosomal conformation changes at schizophrenia-relevant sequences disproportionally occurred in neurons, highlighting the existence of cell type-specific disease risk vulnerabilities in 3D genome organization.
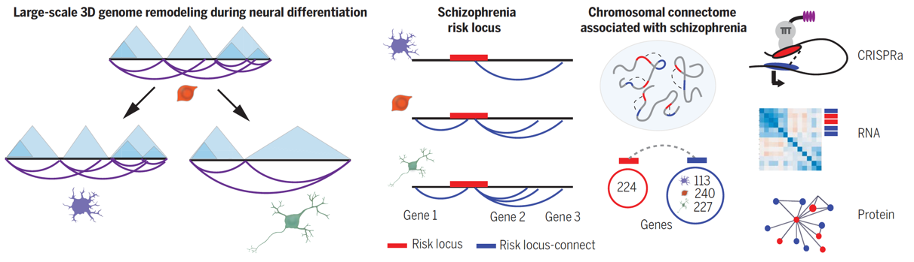
Study design for demonstrating that 3D genome remodeling during neuronal differentiation with parallel expansion of schizophrenia risk space.
Rajarajan, P., et al. (2018). Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science, 362(6420), eaat4311.
Research continues to yield exciting results in this field, with a recent study published in Nature Neuroscience. The 3D genome was assessed in 351 individuals diagnosed with schizophrenia and 739 with bipolar disorder. This work demonstrated that the brains of both schizophrenia and bipolar disorder show coordinated dysregulation of risk-associated regulatory sequences assembled into kilobase- to megabase-scaling chromosomal domains.
Girdhar, K., et al., (2022). Chromatin domain alterations linked to 3D genome organization in a large cohort of schizophrenia and bipolar disorder brains. Nature Neuroscience, 25(4), 474–483.
Expanding beyond psychiatric disease to investigate psychiatric-metabolic comorbidities with 3D genomics has also yielded interesting findings in crucial cell types. For example, Midbrain dopaminergic neurons (MDNs), which mediate cognition, food intake, and metabolism, also play a role in the neurobiological dysfunction of schizophrenia. Investigation of psychiatric and metabolic risk variants in MDNs revealed “Euclidean hot spots” of clustered chromatin domains harboring risk sequences for schizophrenia and elevated Basel metabolic index (BMI).
Espeso-Gil, S., et al., (2020). A chromosomal connectome for psychiatric and metabolic risk variants in adult dopaminergic neurons. Genome Medicine, 12(1), 19.
3D Genomics in Brain Development
In a recent article published in Nature Genetics, Linares-Saldana and coworkers defined a critical role for bromodomain-containing protein 4 (BRD4) in genome organization and embryonic development. Specifically, in the early stages of embryonic development, neural crest differentiation is controlled through 3D genome folding orchestrated by specific genes. Furthermore, their work revealed that BRD4 depletion results in compromised genome folding and loop extrusion, valuable insights for understanding developmental disorders known as cohesinopathies.
Linares-Saldana, R., et al., (2021). BRD4 orchestrates genome folding to promote neural crest differentiation. Nature Genetics, 53(10), 1480–1492.
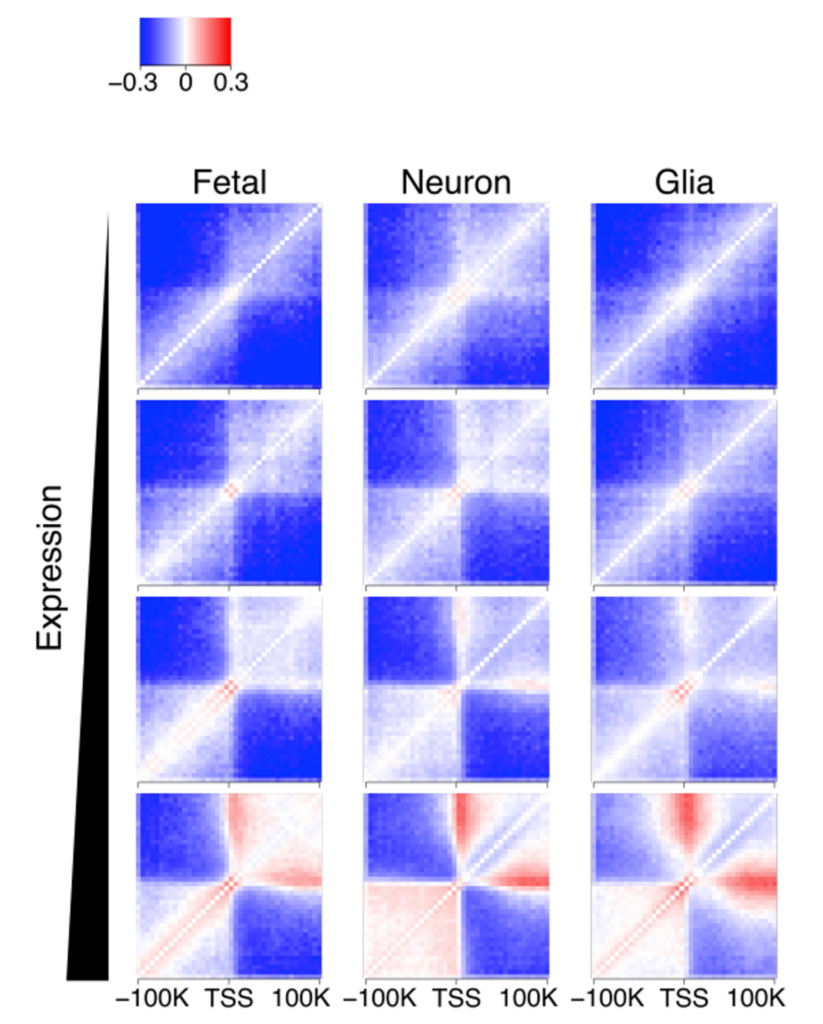
Hi-C interaction heatmap around transcription start sites across 4 different expression levels showing transcription is associated with gene looping.
How the genome is spatially organized into compartments, TADs, and loops can significantly affect gene expression. For example, a bioRxiv preprint led by a group of scientists from the Icahn School of Medicine at Mount Sinai demonstrated that cell type-specific differences between neurons and glia involve broad-scale reorganization of the compartment and TAD structure. In contrast, between neural development and adulthood, changes in the 3D genome primarily involve fine-tuning of regulatory loops.
Rahman, S., et al. (2021). From compartments to gene loops: Functions of the 3D genome in the human brain. bioRxiv Preprint.
In addition to how the brain nervous system develops, the 3D organization of the genome also changes in response to hormonal stimuli and by sex. In a study published in Nature Communications, scientists mapped the 3D genome of ventral hippocampal neurons across the estrus cycle and by sex in mice. In female mice they found cycle-driven dynamism in 3D chromatin organization, including in estrogen response elements-enriched X chromosome compartments, autosomal loops, and enhancer-promoter interactions. This study revealed unique 3D genome dynamics in the female brain relevant to female-specific gene regulation, neuroplasticity, and disease risk.
Rocks, D., et al., (2022). Sex-specific multi-level 3D genome dynamics in the mouse brain. Nature Communications, 13(1), 3438.
Learn more about Arima technology and contact us to explore how you can get started with 3D genomics in your neuroscience research.



