February 13, 2024
Share
The continual evolution of next-generation sequencing technologies allows scientists to sequence more, faster, and at lower costs. At Arima, we strive to ensure that the chemistry behind our HiC kits are compatible with the latest technologies so that our customers can take advantage of their benefits.
Our services department routinely leverages the Illumina NextSeq™ and NovaSeq™ as our sequencing platforms for HiC. To make Arima-HiC+ experiments more widely accessible, we are testing out various sequencers to make it easier for customers to integrate the Arima-HiC+ kits into their existing or preferred sequencing platforms. In our latest efforts, we chose to evaluate the AVITI™ benchtop sequencer from Element Biosciences due to its lower sequencing cost and high base call accuracy.
To assess the potential of the AVITI™ benchtop sequencer from Element Biosciences, we prepared cancer cell lines and solid tumor FFPE tissue samples with the Arima-HiC+ FFPE kit. Following this, we generated sequencing libraries from the HiC samples and sequenced them using both the AVITI™ and the Illumina NextSeq™ sequencers, ensuring equal sequencing depth. Our Arima-SV pipeline then processed the reads for quality assessment, mapping, and structural variant (SV) calling. Key findings from our evaluation are highlighted below, and a detailed technical note is available for download.
High-quality Sequencing Results
Our analysis using the Arima SV Pipeline for Mapping, SV Detection and QC found that the sequencing quality of our samples from the AVITI™ sequencer matched that from the NextSeq™. The only exceptions were that there were slightly fewer % duplicated pairs and slightly higher % unique valid pairs with reads from the AVITI™. This is attributed to the elimination of optical duplicates on the Element AVITI™ platform.
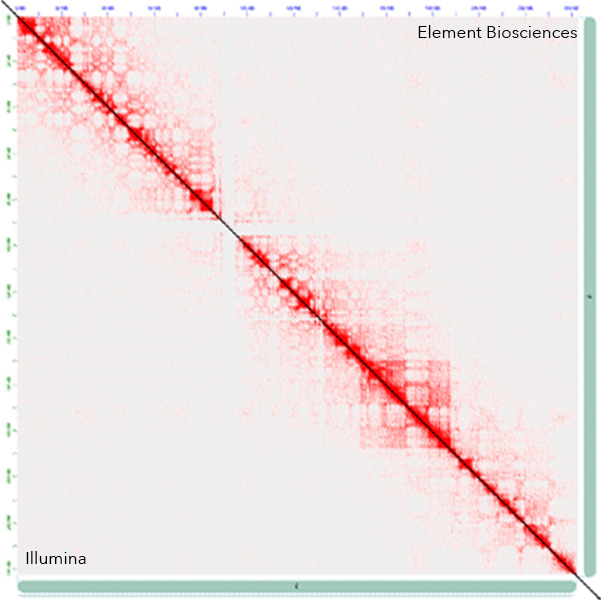
Similar Hi-C Heatmaps
The slight difference in quality mentioned above was reflected in the Hi-C heatmaps generated from the reads. The signal intensity of the heatmaps from the AVITI™ sequencer was slightly higher, likely due to more valid reads from the AVITI™ sequencer. Heatmaps from both sequencers were high-quality and generally agreed with one another.
Structural Variant Calling
We fed the sequencing reads into Arima-SV bioinformatics pipeline to identify rearrangements. In most cases, SVs identified based on the AVITI™ reads were identical to the SVs called based on the NextSeq™ reads. In some cases, we were able to identify SVs from the AVITI™ reads that were not identified from the NextSeq™ reads. We attribute this to the slight increase in the number of unique valid pairs and the slight decrease in percent duplicate pairs from the AVITI™ sequencer vs. NextSeq™.
Based on our evaluation, we can recommend the AVITI™ benchtop sequencer for use with the Arima-HiC+ technology. While this study focused on structural variant calling, our conclusions can be applied to other 3D genome applications (such as calling loops, TADs, and compartments).
We will continue to test our kits with new sequencing technologies to increase options available to our customers. Stay tuned for more updates from our labs!
Download Download the tech note to learn more.
