May 9, 2023
Share
Tumors of the brain and spinal cord, including ependymomas, are the most prevalent tumor type among adolescents 14 years of age and younger in the United States. Although the molecular mechanisms that underlie ependymomas are coming to light, a complete understanding of tumor-driving mechanisms and how to target them for treatment remains elusive. Leading pediatric cancer researchers Lukas Chavez, Konstatin Okonechnikov, and colleagues at the University of California, San Diego, recently uncovered tumor-dependency genes, structural variants (SVs), and pathways characterizing the ependymoma 3D genomic landscape.
Intracranial ependymomas are complex: disease classification is based on anatomical location (supratentorial vs. infratentorial or posterior fossa) and can be further subdivided into distinct molecular groups based on DNA methylation patterns and expression profiling. The most common and aggressive ependymoma molecular groups are the supratentorial ZFTA-fusion associated and the posterior fossa ependymoma group A (PFA).
Recent data have demonstrated that tumorigenesis of ZFTA-fusion associated ependymoma is driven by repeating chromothripsis events that lead to an array of ZFTA-RELA fusion genes. Conversely, the absence of recurrent mutations or gene fusions suggests that PFA tumors may be epigenetically driven. However, developing research has yet to discover small molecules that can target gene fusions or epigenetic pathways. Therefore, further investigation into the 3D genome of both ZFTA and PFA ependymomas is required.
Chavez and colleagues thoroughly examined the 3D chromatin architecture of ZFTA and PFA ependymomas from frozen and FFPE tumor biopsy samples by leveraging the Arima-HiC+ FFPE Kit and Arima promoter capture HiC kit together with a multi-omics approach including, ChIP-seq, gene expression, and DNA methylation analyses. Recently published in Nature Communications, their work provides new insights into targetable genetic alterations and potential therapeutic options for pediatric cancers.
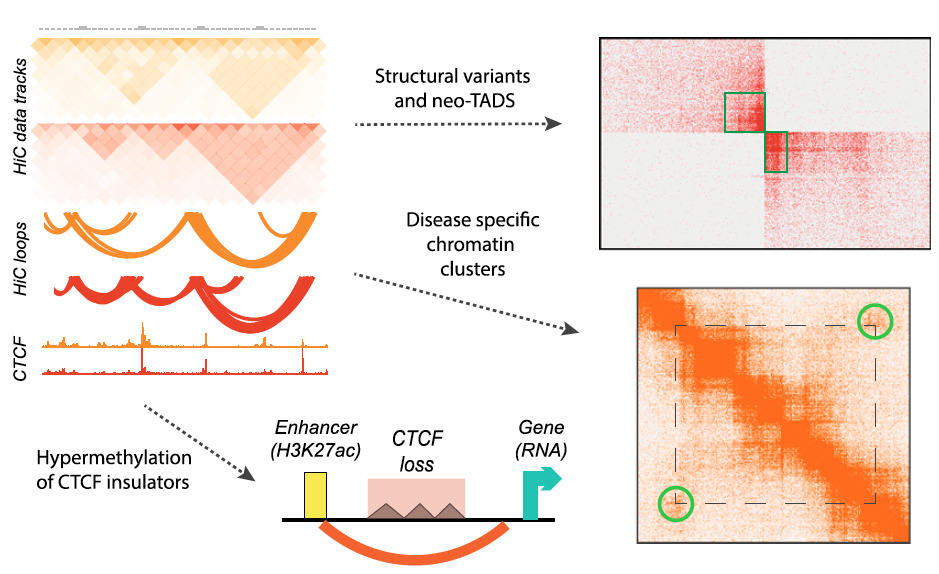
Summary of the major results obtained using genome-wide Hi-C in ependymoma brain tumors (Okonechnikov et al., 2023).
Using 3D Genomics to Identify Challenging Genetic Alterations in Ependymoma – Highlights
- Hi-C data from fresh frozen or FFPE biopsy samples stratified ependymoma brain tumors into distinct group-specific 3D tumor genome conformations. Computational analyses of SVs, a first for frozen and FFPE samples from ependymoma patients, reliably detected SVs in ZFTA and RELA gene loci in supratentorial ZFTA but not in PFA tumors. These Hi-C data also captured complex rearrangements at chromosome 11 in some, but not all, ZFTA ependymoma tumor samples, suggesting inter-chromosomal rearrangements beyond chromosome 11.
- The generation of new topologically associating domains (neo-TADs) plays a critical role in oncogenesis. Computational reconstruction of the tumor genome revealed neo-TAD formation in all ZFTA samples and implicated the REST Corepressor 2 (RCOR2) gene as a key player in ZFTA-RELA In patient-derived cell-lines, RCOR2 shRNA-mediated knockdown reduced ZFTA cell growth but had less of an impact on PFA cells.
- In all PFA samples, Hi-C data revealed a 3D chromatin cluster located more than 4 million base pairs apart on chromosome 2 with no sign of similar DNA interactions in ZFTA These data suggest that this chromatin cluster is potentially unique to the cellular lineage of PFA ependymoma.
- Hi-C analyses in PFA samples revealed complex intra- and inter-chromosomal rearrangements and characteristic 3D chromatin organizations. These data further uncovered epigenetic integrin signals (ITGA6 and LAMC1) as a potential mechanism that exacerbates cell proliferation.
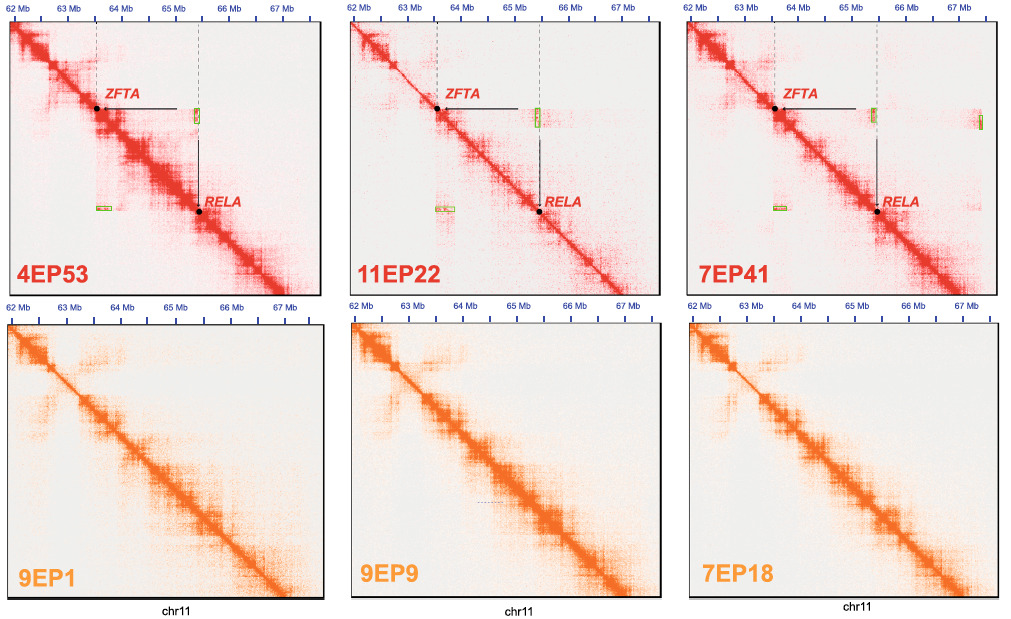
Hi-C data detects structural variants that drive ZFTA-RELA fusion genes only in supratentorial ZFTA-fusion associated tumors (top row) but not in PFA tumors (bottom row) (Okonechnikov et al., 2023).
The authors emphasize that “overall, our study has identified several group-specific tumor dependencies in ependymomas, opening avenues for potential therapeutic interventions that are urgently needed for this disease, especially for ZFTA and PFA ependymoma patients. We anticipate that our results will lay the foundation for further preclinical validation experiments in ependymoma patient derived xenografts (PDX)45 or in other in vivo models such as mouse models with ZFTA-RELA fusions.” They also note that their study “shows that the analysis of 3D tumor genome may also be relevant for other (pediatric) cancers, for which the drivers are known but there are few therapeutic options.”
Learn more about 3D genomics for cancer research or request a project consultation.
Resources
Okonechnikov, K., et al. (2023). 3D genome mapping identifies subgroup-specific chromosome conformations and tumor-dependency genes in ependymoma. Nature Communications, 14, 2300.



