Generate Comprehensive Insights
Generate dependable results from FFPE tissue samples to detect both coding and non-coding structural variants in this hard-to-use sample type. Learn more about our structural genomics application.
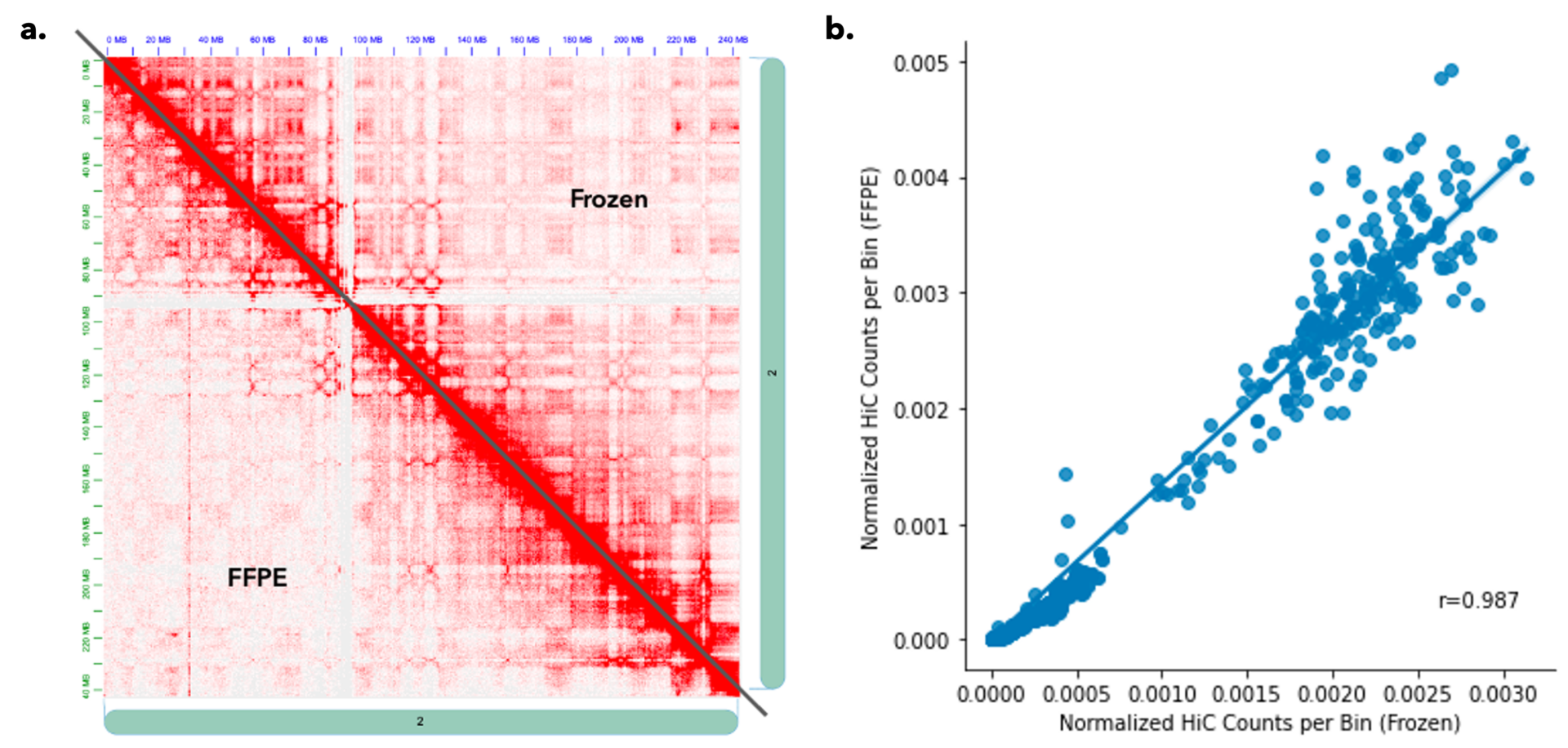
FFPE Sample Results are Highly Correlated to Frozen Tissues. Matched frozen and FFPE tissue of K562 cell cultures were processed with the Arima-HiC+ and Arima-HiC+ FFPE kits, respectively. Libraries were generated following Arima protocols and sequenced using Illumina next generation sequencing (57M and 27M reads were generated, respectively). Representative data for Chromosome 2 at 500 Mb are shown in a. Hi-C contact heat map (500Mb resolution) and b. correlation plot of the normalized Hi-C counts per bin for frozen versus FFPE samples showing a high correlation (r=0.987).
Arima SV Detection Assay Workflow
An end-to-end solution to take you from sample to insights.
Sample Prep
Arima-HiC+ / Arima-HiC+ FFPE Chemistry
Works with cells, liquid biopsy, fresh/frozen tissue and formalin-fixed, paraffin embedded (FFPE) tissue.
Library Prep
Arima Library Prep
Sequencing
Next Generation Sequencing
2 x 150 paired-end reads
Tested on Illumina, Element and Singular NGS platforms.
Data Analysis
Arima Bioinformatics Platform
Leverage the Arima SV QC and Deep Sequencing pipelines.
Compatible with 50–500 million PE reads / clusters. We recommend at least 100M reads.
Matija Snuderl, MD
Director of Molecular Pathology and Diagnostics, NYU Langone Medical CenterExplore Our Resources
Learn more about how you can use Arima HiC+ Kits and Workflows in your research: