November 3, 2021
Share
Uncovering new insights into an oncogenic driver that has been highly studied for nearly 30 years requires a fresh approach. Dr. Stephen Lessnick, professor of pediatrics at The Ohio State University College of Medicine, has spent his research career studying EWS/FLI, the driver mutation for Ewing sarcoma.
To gain a new perspective, Dr. Lessnick and his team at Nationwide Children’s Hospital, including Dr. Emily R. Theisen (@TheisenLabNCH), decided to apply an unbiased whole-genome approach. With in situ Hi-C they were able to precisely define the global changes in chromatin structure mediated by EWS/FLI in Ewing sarcoma and further link these structural changes to alterations in gene expression.
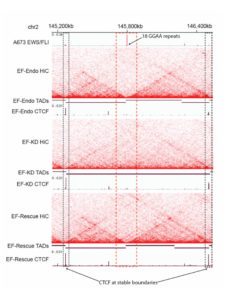
EWS/FLI enriched in Ewing sarcoma cell TAD boundaries. Heatmap indicating Hi-C interactions in Ewing sarcoma cell lines, revealing enrichment of EWS/FLI in TAD boundaries in Ewing sarcoma cells (top and bottom), but absent in the knockdown line (middle).
Ewing sarcoma is a highly aggressive pediatric cancer, causing bone tumors in children and adolescents. Genetically, Ewing sarcomas are characterized by chromosomal translocations in which a member of the FET gene family is fused with an ETS transcription factor. EWS-FLI, a chimeric transcription factor present in 85% of cases, results in reprogramming of the epigenome by inducing de novo enhancers at GGAA microsatellites. Ultimately, this reprogramming results in dysregulation of thousands of downstream targets and oncogenesis.
Despite proven effectiveness for treatment of localized disease, long-term survival for patients with metastatic or relapsed Ewing sarcoma remains low, underscoring the need to further elucidate how changes in the epigenome impact Ewing sarcoma initiation and progression.
Using the Arima HiC+ Kit to explore genome-wide 3D chromatin conformation data, Dr. Lessnick’s team was able to fill significant knowledge gaps in Ewing sarcoma, which they published in this bioRxiv preprint
Using 3D Genomics to Understand Ewing Sarcoma – Highlights
- EWS/FLI was significantly enriched in topologically associated domain (TAD) boundaries in Ewing sarcoma cells, suggesting a direct involvement for EWS/FLI in altering TAD boundaries.
- EWS/FLI bound to GGAA microsatellites has a direct role in anchoring chromatin looping (both inter- and intra-TAD loops) and TAD boundaries, as well as promoting active compartmentalization of chromatin.
- While putative EWS/FLI target genes have been identified previously, no study to date has been able to probe genome-wide chromatin loop data associated with EWS/FLI to accurately link target genes to specific EWS/FLI bound regulatory elements. The team identified “direct EWS/FLI” target genes with transcription start sites within 20 kb of an EWS/FLI binding site, as well as differential expression in Ewing sarcoma cell lines
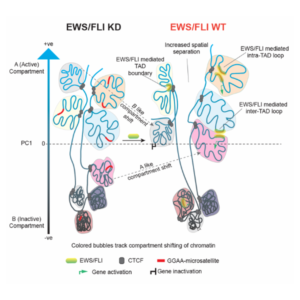
Model illustrating EWS/FLI chromatin changes. High-resolution analysis of 3D chromatin isolated from patient-derived Ewing sarcoma cells reveals reprogramming of higher order chromatin interactions, changes in A/B compartments, TAD boundaries and enhancer-promoter chromatin loops. This reprogramming of the genome ultimately facilitates an oncogenic transcriptional state in Ewing sarcoma.
The authors note that “These data demonstrate a key role of EWS/FLI in mediating genome-wide changes in chromatin configuration and support the notion that fusion transcription factors serve as master regulators through three-dimensional reprogramming of chromatin.” It is likely that similar fusion oncoproteins found in different cancers are involved in reorganization of chromatin to promote oncogenesis. For example, a recent study in Nature similarly showed that an oncogenic fusion transcription factor (NUP89-HOXA9) can promote chromatin looping to regulate gene expression and oncogenesis.
These and other novel insights into altered transcriptional states in cancer have been identified using Arima Hi-C technology to explore the 3D chromatin landscape.
Learn more about 3D genomics or request a project consultation.
Resources
Showpnil, I. A. et al. (2021) EWS/FLI mediated reprogramming of 3D chromatin promotes an altered transcriptional state in Ewing sarcoma. bioRxiv 2021.09.30.462658; doi: https://doi.org/10.1101/2021.09.30.462658



